| PI | Institution | Title |
|---|---|---|
| Jonathan Oliner | Affymetrix | Reverse-Engineering Signal Transduction Networks |
The CustomSeq array was one of the first to offer researchers a powerful DNA analysis tool on the same proven Affymetrix platform that has since become the industry standard for mRNA gene expression research. CustomSeq arrays are based on GeneChip technology that have set a standard in large-scale resequencing performance. Applications include candidate gene studies, pharmacogenetic studies of genetic variation across patients in clinical trials and pathogen subtyping to identify known and novel strains of microorganisms. Affymetrix' resequencing array offers researchers numerous benefits, including the following:
- CustomSeq arrays deliver completed sequence that is greater than 99.99 percent accurate in just 48 hours, with minimal hand curation or sequence alignment. It can take weeks to get completed sequence using capillary sequencing methods.
- CustomSeq array content is determined by the researcher, who can choose any combination of contiguous genomic regions or dispersed sets of exons.
- Affymetrix' parallel manufacturing method and stringent quality control ensures 99.9999% reproducibility between CustomSeq arrays.
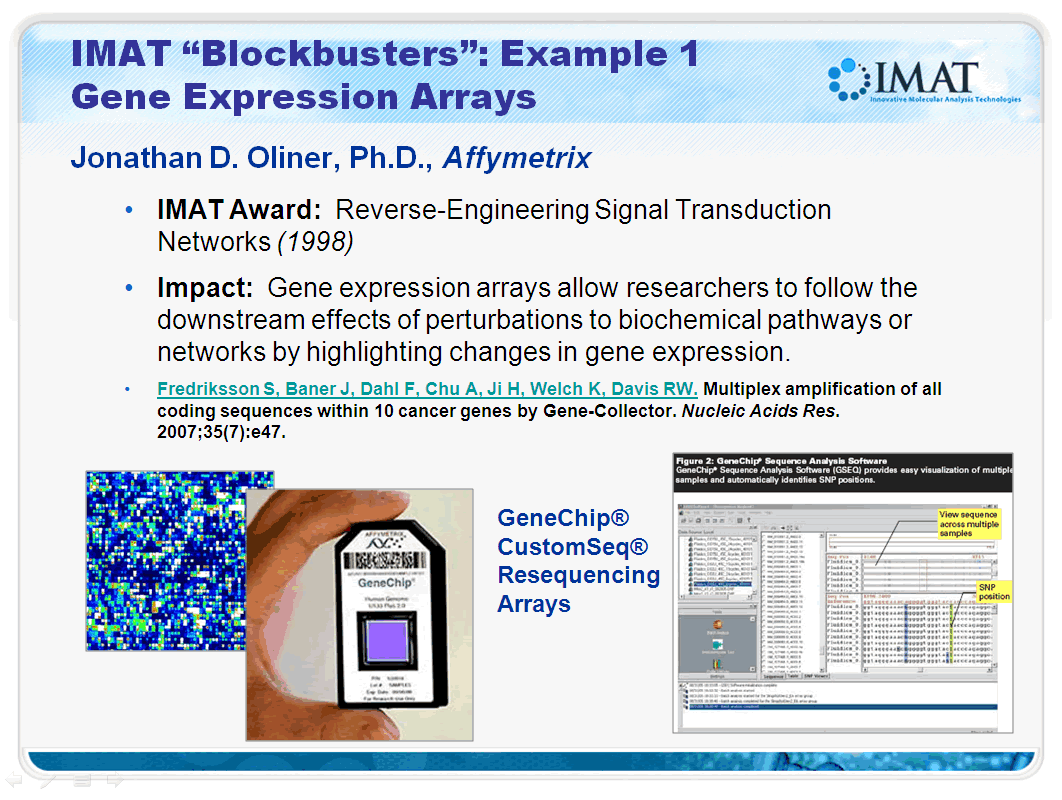
IMAT "Blockbusters": Example 1 Gene Expression Arrays
Jonathan D. Oliner, Ph.D., Affymetrix
- IMAT Award: Reverse-Engineering Signal Transduction Networks (1998)
- Impact: Gene expression arrays allow researchers to follow the downstream effects of perturbations to biochemical pathways or networks by highlighting changes in gene expression.
- Fredriksson S, Baner J, Dahl F, Chu A, Ji H, Welch K, Davis RW. Multiplex amplification of all coding sequences within 10 cancer genes by Gene-Collector. Nucleic Acids Res. 2007;35(7):e47.

